ExerGeneDB Project Overview
All data from the ExerGeneDB study were collected from public databases, including NCBI and NGDC. We made every effort to thoroughly search for data related to physical exercise and relevant information. The collected information can be categorized into three parts: Sequencing, Sample, and Exercise Information. Based on this information, we meticulously organized all the data and conducted a Differential Expression Gene (DEG) analysis.
Basic Information
The information within the ExerGeneDB project is categorized into three main parts: Sequencing, Sample, and Exercise Information. Notably, the information for mice and humans shares a fundamental similarity. However, it is crucial to acknowledge that human exercise patterns exhibit a higher level of complexity compared to mice. In light of this, ExerGeneDB has established distinct exercise patterns specifically tailored for humans. It is worth noting that certain gender-related information is missing but can be inferred. To address this gap, a machine learning model is employed to deduce unknown gender information based on sex-related genes.
Sequencing informationSequencing type
Sequencing technology
| Sample information
| Exercise information
|
Organism: Mus musculus and Rattus norvegicus
Sequencing informationSequencing type
Sequencing technology
| Sample information
| Exercise information
|
Organism: Homo sapiens
Organ/Tissue/Cell Type
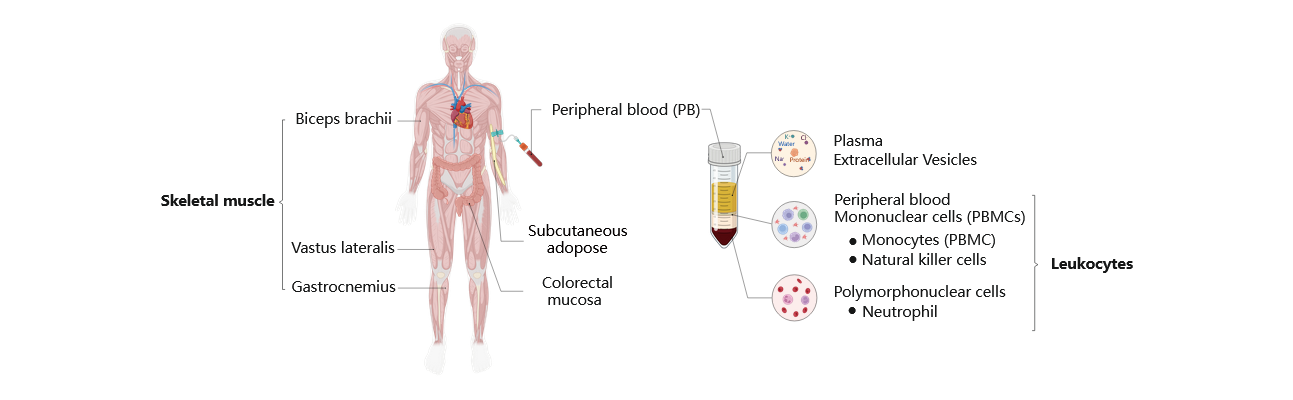
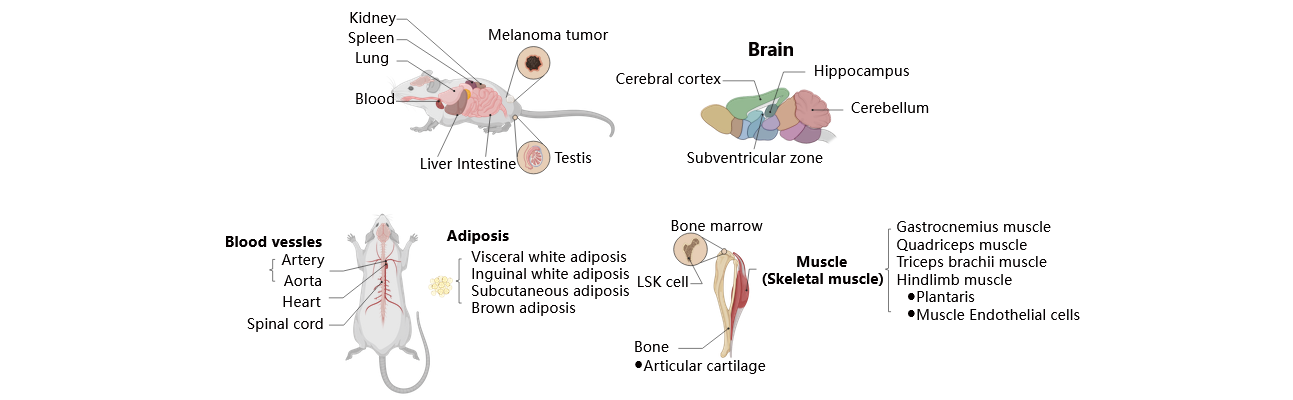
The ExerGeneDB project extensively collects high-throughput sequencing data from various tissues and organs. Detailed information on these tissues/organs is provided in the accompanying images. However, certain data, particularly in bulk RNA-seq, lacks clear sampling site details. For instance, some papers only specify their data source as ‘muscle’ without specifying the specific type of muscle. In order to establish a standardized and organized database, we have introduced an additional two-column information system to address unclear details. For instance, the tissue information in ExerGeneDB now consists of two columns. The first column retains the original paper's information, while the second column provides clarification defined by ExerGeneDB. This approach aims to enhance the clarity and completeness of the dataset.
Homo sapiens
Mus musculus / Rattus norvegicus
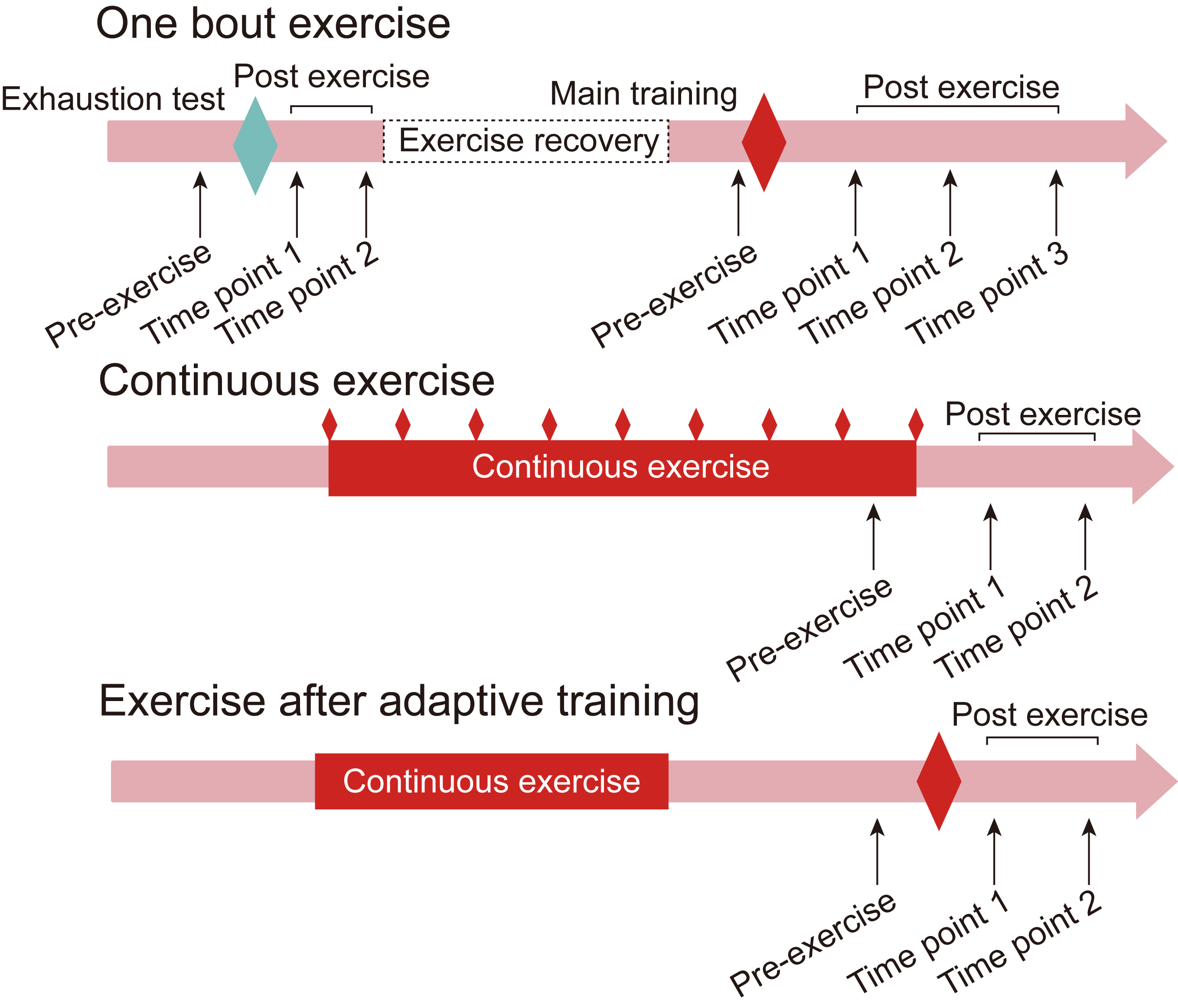
Exercise Pattern (Homo sapiens)
The exercise patterns for mice/rats are generally simple, whereas in humans, exercise protocols are often intricate and multifaceted. ExerGeneDB categorizes all clinical RNA-seq data and classifies them into three distinct exercise patterns. ExerGeneDB defines the first exercise pattern as ‘One bout exercise’, where participants undergo a single main training session, and tissues are subsequently collected. However, the actual scenarios can be more intricate, as participants may undergo multiple sessions in a single day, or they might be required to undergo an exhaustion test several weeks before the main training. The second exercise type is termed ‘Continuous exercise’, involving participants engaging in sustained physical activity over several days, weeks, or months. The last pattern is ‘Exercise after adaptive training’, signifying that participants have undergone a training period before participating in a main training session. This nuanced classification aims to capture the diverse nature of human exercise routines within the ExerGeneDB framework.
Exercise Intensity
ExerGeneDB has redefined various levels of exercise intensity based on information extracted from original articles.
Mus musculus/Rattus norvegicus | Homo sapiens |
Voluntary wheel running Low-intensity: 2~5 km/day Moderate-intensity: 5~11 km/day High-intensity: 11~ km/day Treadmill running Low-intensity: 0~10 m/min, 30~120 min/day, 0 degree Moderate-intensity:11~22 m/min, 30~120 min/day, 0 degree High-intensity: 22~ m/min, 30~120 min/day, 0 degree Swim exercise Moderate-intensity: 30~60 min/time, 3~5 times/week High-intensity: 90~ min, 5~ times/week VO2max Low-intensity: 0~50% VO2max Moderate-intensity: 50%~70% VO2max High-intensity: 71%~90% VO2max | Aerobic exercise VO2max Low-intensity: 0~50% VO2max Moderate-intensity : 51%~75% VO2max High-intensity:75%~ VO2max MET Low-intensity:1.5~3 MET Moderate-intensity : 3~4MET(female) 3~6MET(male) High-intensity: 4~ MET (female) 6~ MET (male) HRMax Low-intensity: 0~60% HRMax Moderate-intensity: 61%~70% HRMax High-intensity: 71%~85% HRMax Resistance exercise RM Low-intensity RT: 0~50 %1RM Moderate-intensity RT: 51%~69% 1RM High-intensity: 70%~ 1RM |
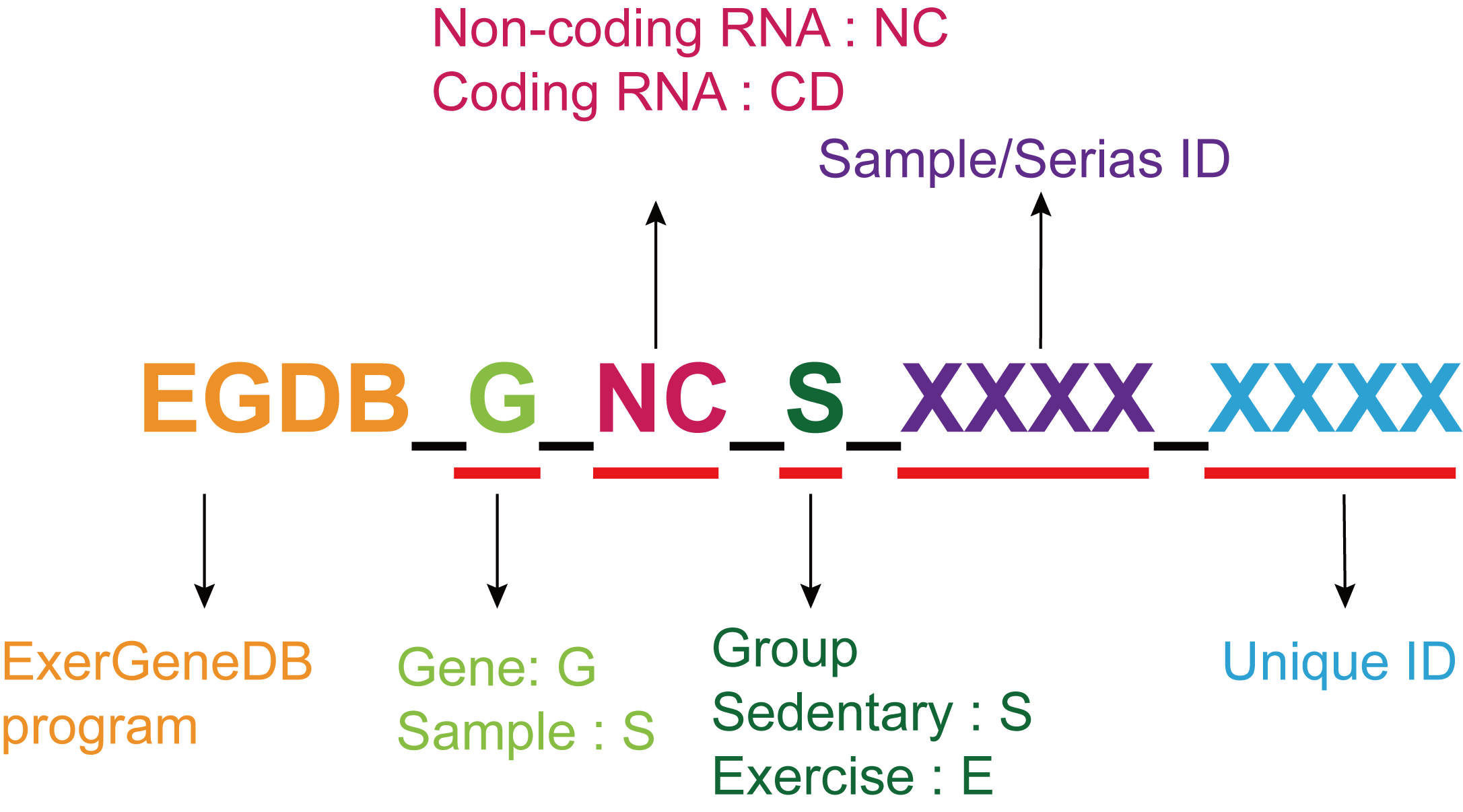
ExerGeneDB ID
For the sake of efficient database management, we have assigned a unique identifier, EGDB_G_NC_S_XXXX_XXXX, to each entry in our database, which we refer to as the "EGDB ID". This EGDB ID serves as the unique identifier for every entry in the database. It comprises six parts: firstly, the ‘Project Name’ – our project is called ExerGeneDB, abbreviated as EGDB; secondly, the ‘name of the gene or sample’ – genes are denoted by G and samples by S; next is the ‘RNA type’ – coded RNA is abbreviated as CD, non-coding RNA as NC, and genes originating from single-cell data as SC. Of course, if the EGDB ID is used to represent a sample, this part is omitted. Subsequently, there's the ‘sample/dataset name’ – for gene differential data, this column displays the digits from the dataset number, while for sample information, it displays the digits from the sample number. Lastly, there's the unique identifier code. To ensure that the preceding information is sufficient for unique identification, we introduce a series of digits to transform the EGDB ID into a unique identifier code.