Welcome to ExerGeneDB!
ExerGeneDB provides users with an interactive query function for transcriptome data, it consists of two main functional modules: 'Search' and 'Explore'.
This tutorial aims to provide a comprehensive understanding of the two distinct modules offered by ExerGeneDB, enabling users to effectively utilize the database and access desired information through targeted gene queries.
Search Module
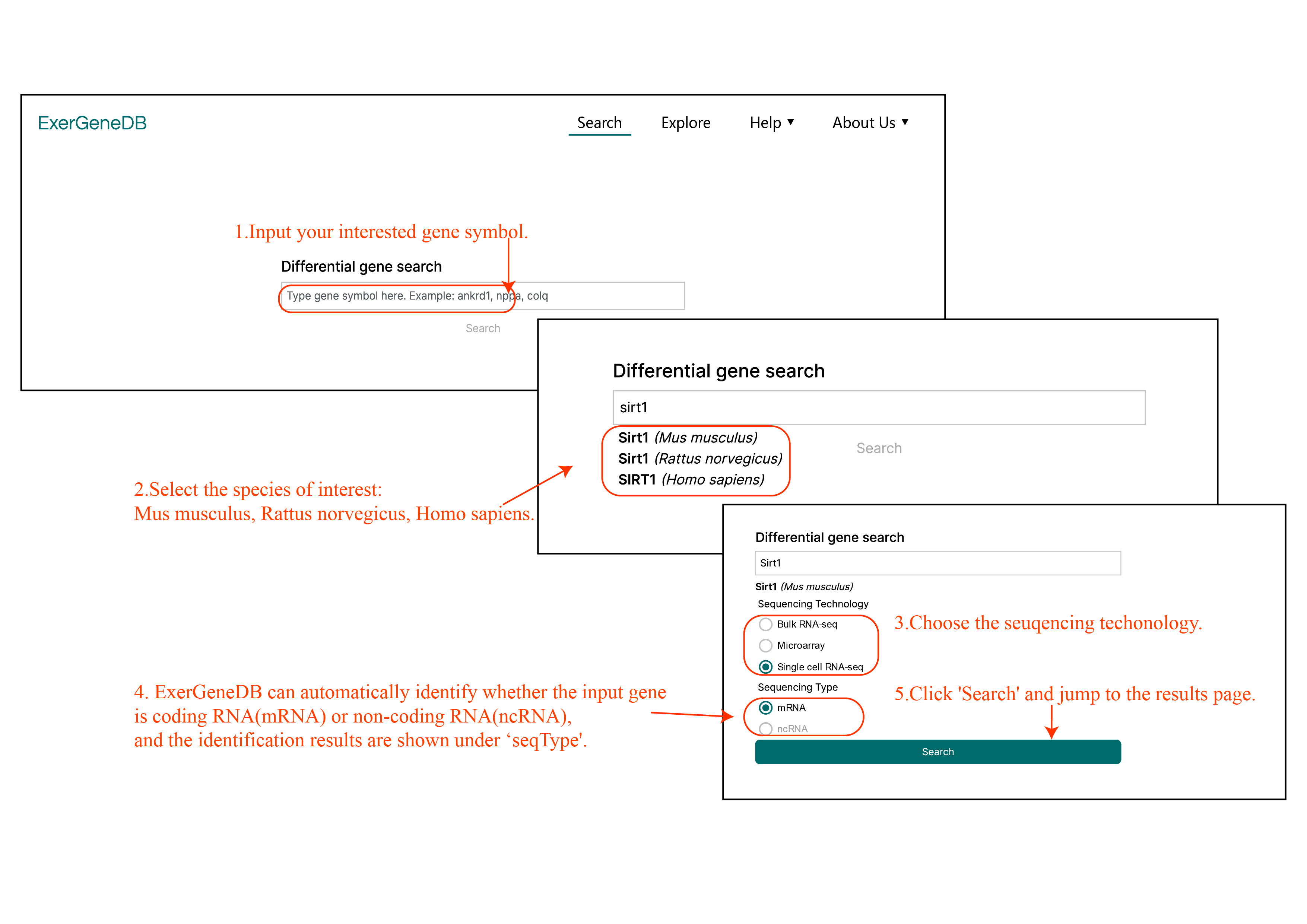
The 'Search' page is designed for querying genes of interest.
Firstly, users can input the gene symbol which they interest into the search box (Tutorial figure 1).
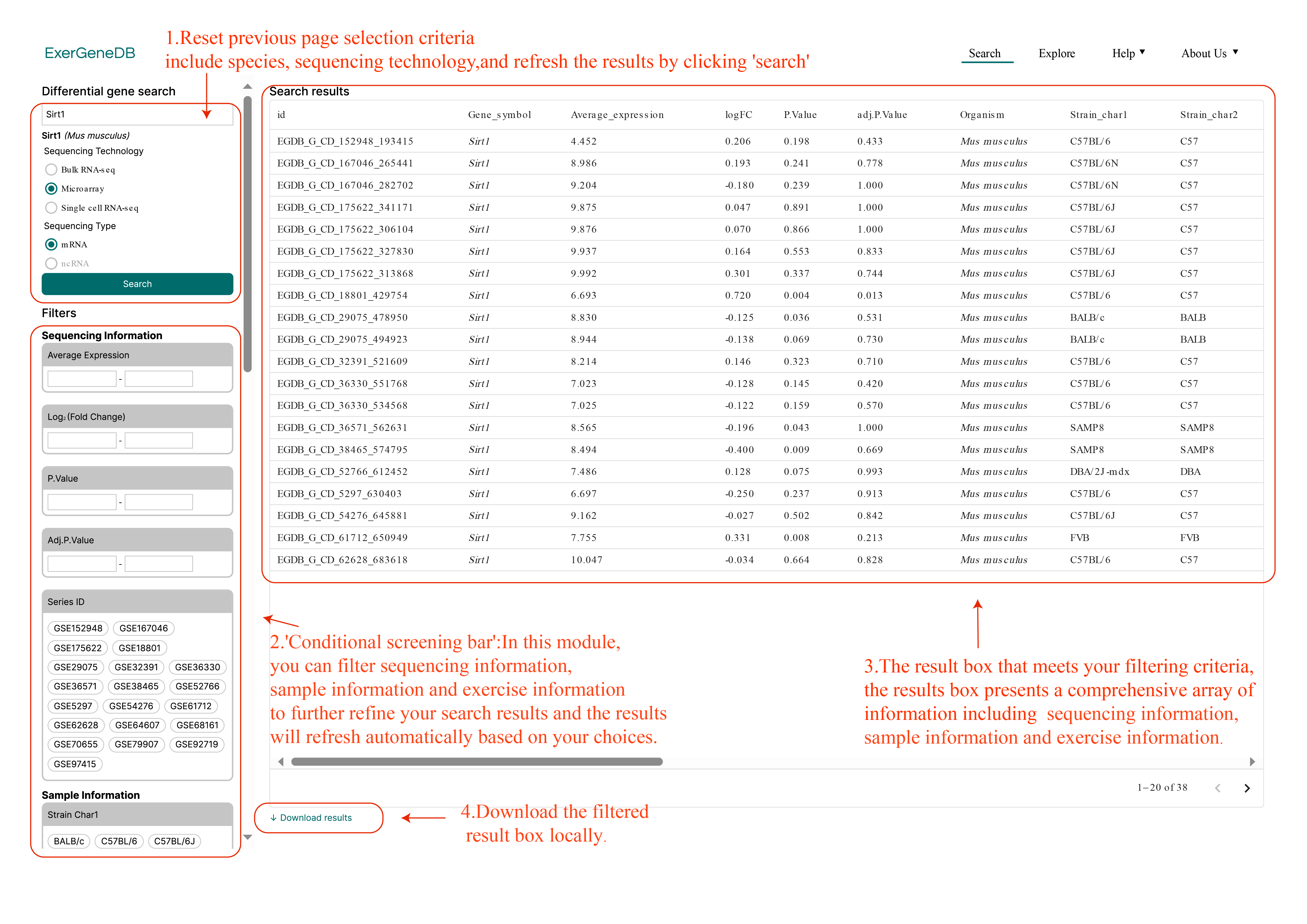
In the results page, users can filter sample, exercise, and sequencing information using the "conditional screening bar" on the right side of the page. At the same time, users can also re-select the information selected on the previous page (Tutorial figure 2).
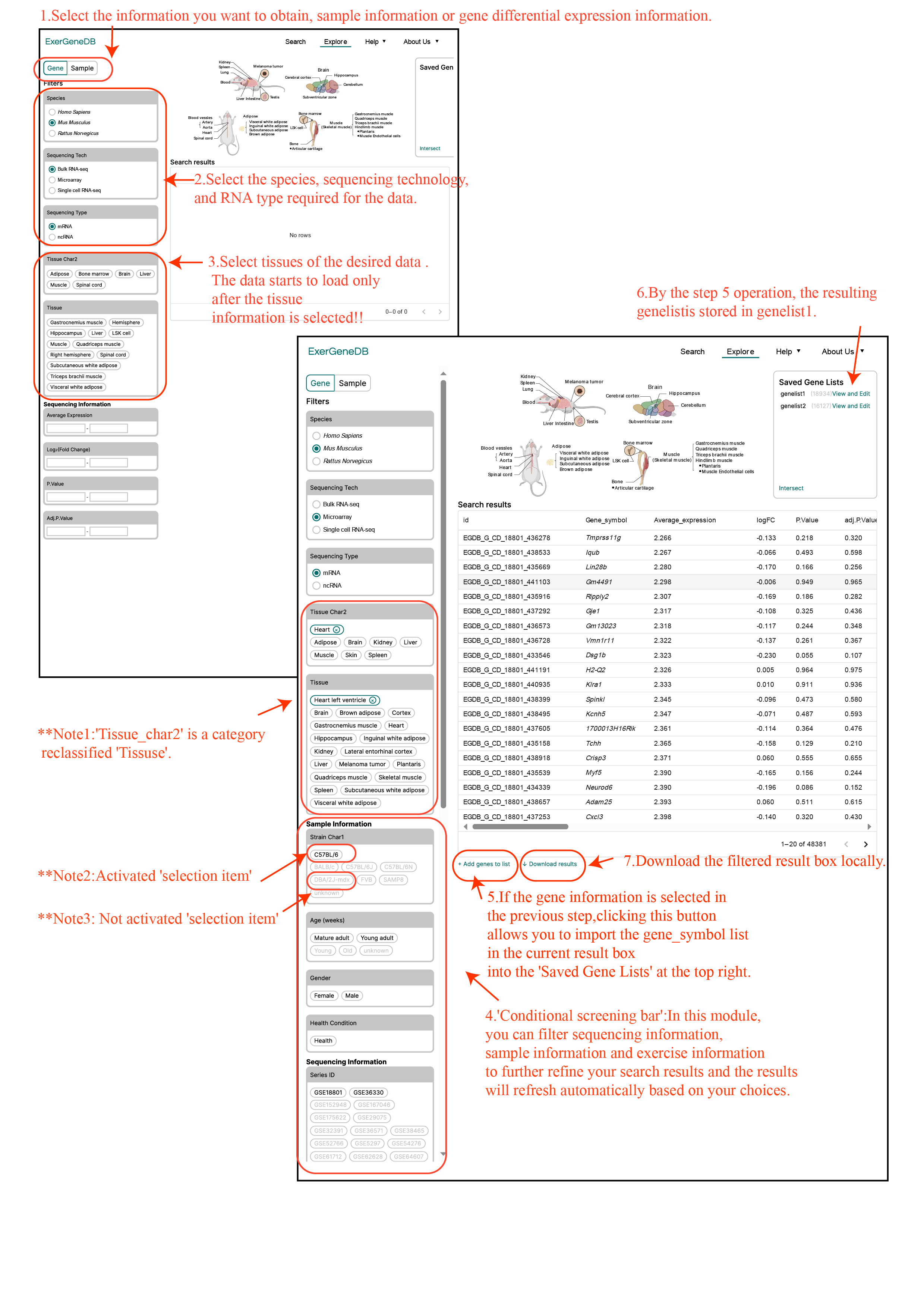
Explore Module
The 'Explore' page enables users to retrieve DEGs or samples for specific organs under particular exercise condition.
For instance, users can utilize the "conditional screening bar" located on the right side to specify their desired tissue and exercise mode, enabling them to retrieve either differential genes or samples. Following this, users can further refine their search by selecting species information, sequencing technology, and RNA type. Once these criteria are set, the database will fetch all relevant data and populate the 'Conditional screening bar'.
The activation of ‘selection item’ in the 'Conditional screening bar' depends on tissue selection. If there is no corresponding data for a selected item in the chosen tissue, the ‘selection item’ will be inactive and appear grayed out. It should be noted that selecting certain options may deactivate others.
Additionally, if users wish to explore the communication of DEGs under various conditions, they can choose to click "Add gene" at the bottom of the result bar to temporarily store the currently screened results in the 'gene list' (Tutorial figure 3). The meaning of the column name in the result box are showing in Tutorial table 1.
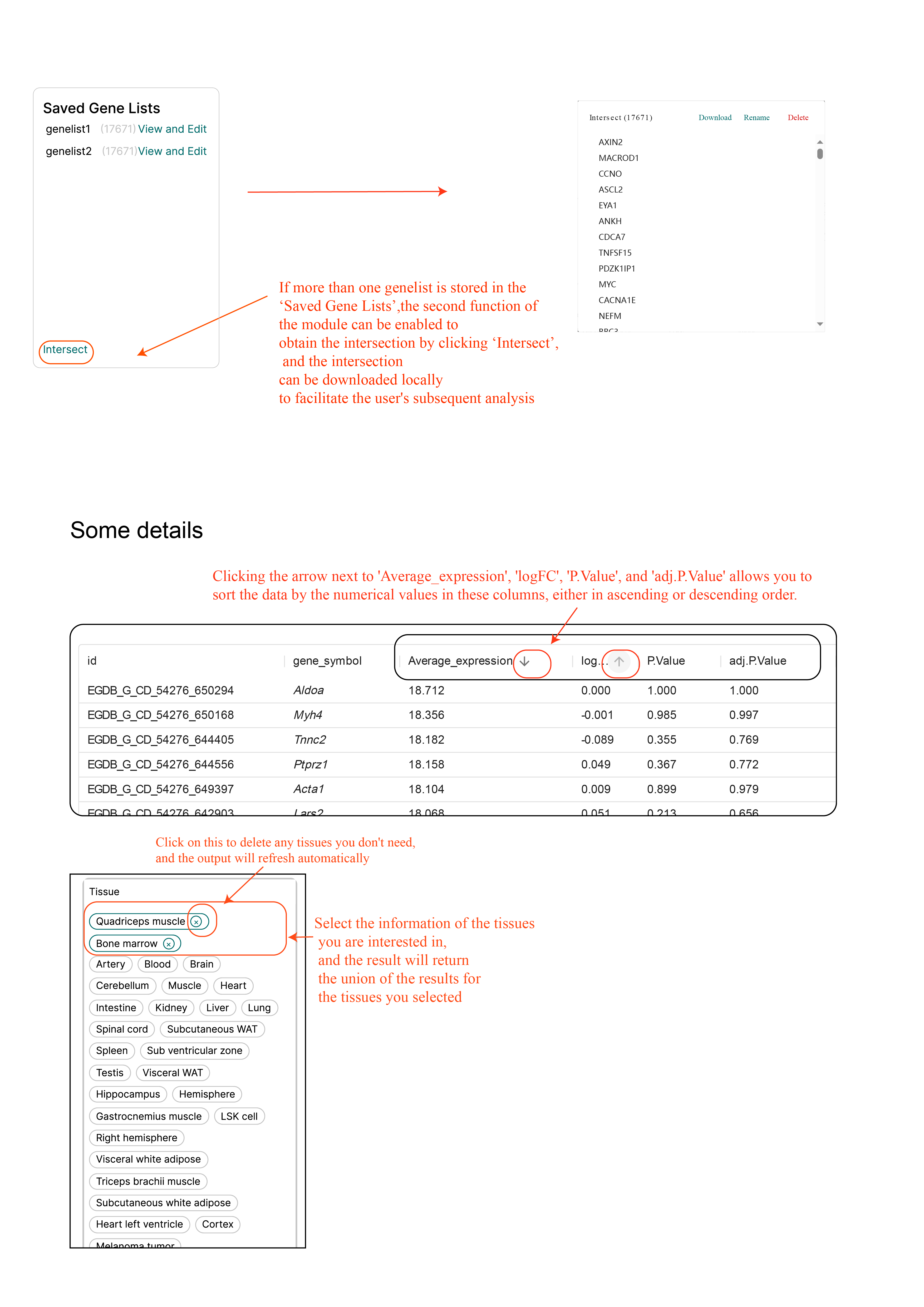
When multiple filter results are in the 'gene list' at the same time, ExerGeneDB supports the intersection of these filter results. Subsequently, users can conduct GO enrichment analysis on the gene sets after intersecting and export the results (The GO enrichment function will online in the nearly future). At the same time, some details are suggested in the diagram below, such as how to choose the organization, the order of the values (Tutorial figure 4).
Tutorial table 1
| Category | Species | Term | Description |
|---|---|---|---|
| Sequencing information | Common | Average_expression | The average of gene expression in "unit dataset". |
| logFC P. Value adj. P. Value | Differential expression between pre- and post-exercise in the 'unit dataset'. Additionally, P values and adj. P values are provided to assess the reliability of the data. | ||
| Seqencing technology | Sequencing technology includs Bulk RNA-seq and single RNA-seq (Micro-array sequencing and High-throughput RNA sequencing) | ||
| RNA type (Sequencing type) | Coding RNA (mRNA) and Non-coding RNA (ncRNA). | ||
| Series_id | Dataset's original ID | ||
| Clinical information | Common | Organism | Homo Sapiens, Mus Musculus and Rattus Norvegicus |
| Tissue_chr1 | “Tissue_chr1” indicates samples sampling location and some of them are precise located to specific organs,tissues and cell types. | ||
| Tissue_chr2 | “Tissue_char2" indicates the organ or tissue source. | ||
| Health_condition | The health state of a sample. | ||
| Human | Age | Human: Minor: 0-18 years old; Young adults: 18-37 years old; Middle aged: 37-65 years old; Elderly: 65 years old and above. | |
| Gender | Male or female | ||
| BMI | Body mass index (BMI) is calculated by dividing a person's weight in kilograms by the square of their height in meters (kg/m2). BMI: Normal:18.5-24.9; Overweight: 25-29.9; Obesity: 30 and above. | ||
| Mouse/Rat | Strain_char1 | The strains of Mus Musculus /Rattus Norvegicus | |
| Strain_char2 | Partition of “Strain_char1” | ||
| Age | Mouse/Rat: Young: 0-8 weeks; Young adults: 8-12 weeks; Mature adult: 12-24 weeks; Middle aged: 40-63 weeks; Elderly: 64 weeks and above. | ||
| Gender | Male or female | ||
| Exercise information | Common | Exercise_time | Frequency of exercise |
| Exercise_frequency | Frequency of exercise | ||
| Exercise_during | Total duration of exercise | ||
| Exercise_intensity | Exercise intensity included low intensity, moderate intensity and high intensity | ||
| Human | Exercise_ type | Human's primary types of exercise including aerobic exercise, resistance training, and combined training. | |
| Exercise_ pattern | ExerGeneDB defines three exercise patterns, the first exercise pattern as 'One bout exercise,' where participants undergo a single main training session, and tissues are subsequently collected. However, the actual scenarios can be more intricate, as participants may undergo multiple sessions in a single day, or they might be required to undergo an exhaustion test several weeks before the main training. The second exercise type is termed 'Continuous exercise,' involving participants engaging in sustained physical activity over several days, weeks, or months. The last pattern is 'Exercise after adaptive training,' signifying that participants have undergone a training period before participating in a main training session. | ||
| Exercise_modality | Exercise modalities of human include walking, running, cycling, and muscle strength training. Additionally, the database includes two other exercise modalities: combined training which combines resistance exercise with one or more aerobic exercises, and aerobic endurance training which involves a combination of more than two aerobic exercise modalities. | ||
| Mouse/Rat | Exercise_modality | The Exercise modalities of mouse/rat includs voluntary wheel running, Swimming exercise and treadmill running. |
Note: ExerGeneDB introduces a concept of 'unit dataset'. ExerGeneDB selects sample datasets with identical sequencing, clinical, and exercise information as units of analysis, which is defined as 'unit dataset'. In a selected unit dataset, samples post-exercise are designated as the experimental group, whereas pre-exercise samples serve as the control group for differential analysis.
Consequently, the information associated with each gene in the results encompasses not only its individual changes but also the characteristics of the 'unit dataset' in which the gene is situated.